 Fig. 1.
Fig. 1.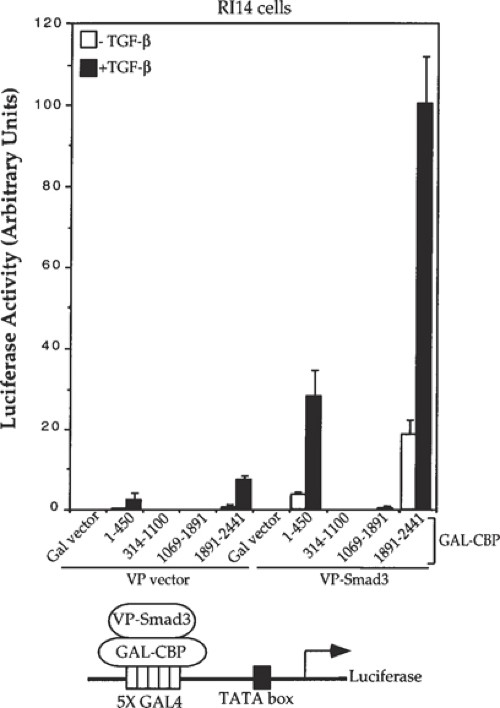 Fig. 2.
Fig. 2.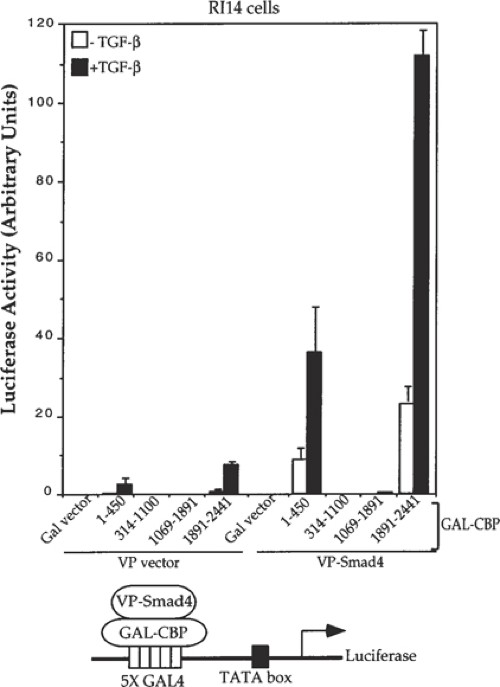 Fig. 3.
Fig. 3.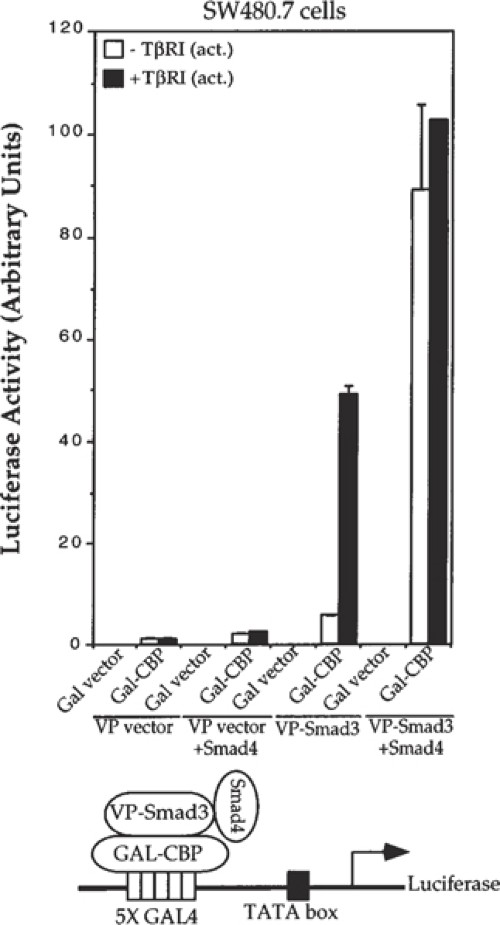 Fig. 4.
Fig. 4.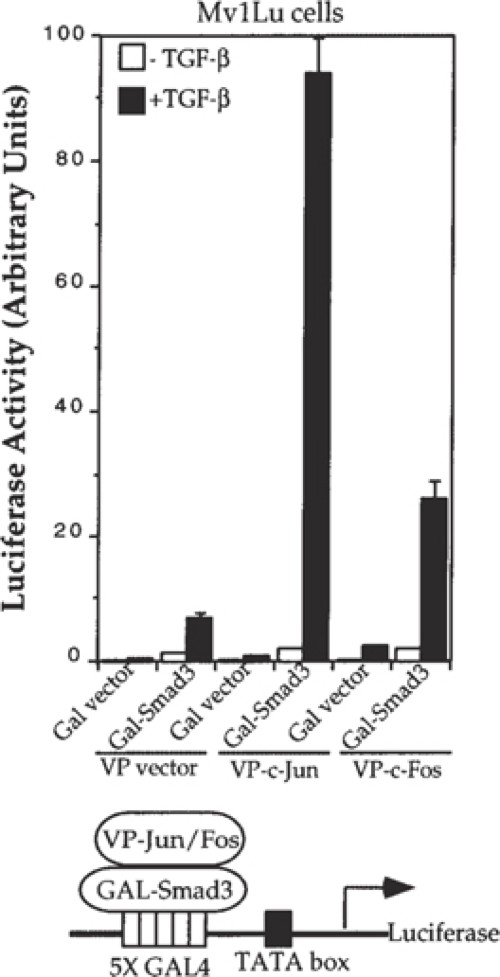 Fig. 5.
Fig. 5.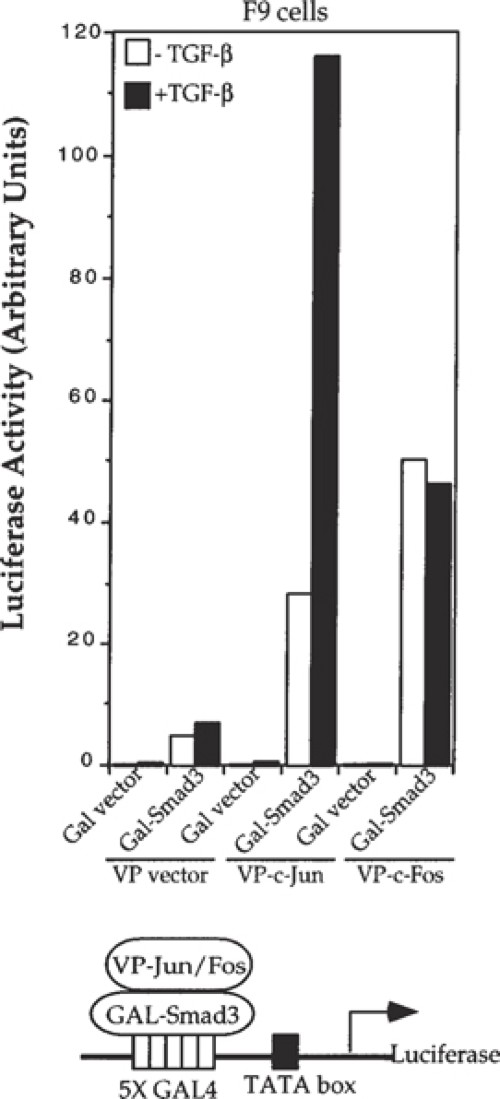 Fig. 6.
Fig. 6.Mammalian two-hybrid assays are used to analyze protein-protein interactions in transiently transfected mammalian cells. The concepts and principles that form the basis for mammalian two-hybrid assays are essentially the same as for the yeast
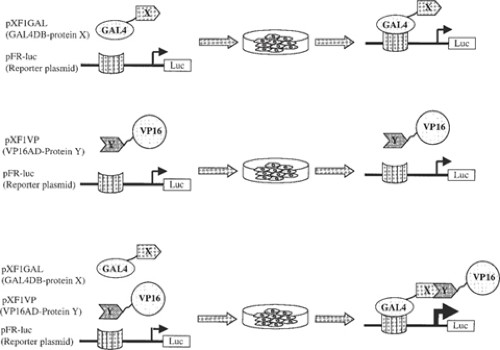
Mammalian two-hybrid assays are used to analyze protein-protein interactions in transiently transfected mammalian cells. The concepts and principles that form the basis for mammalian two-hybrid assays are essentially the same as for the yeast two-hybrid assays (1), discussed in other chapters, except that the analyses are carried out in transfected mammalian cells (2). Thus, similar to yeast two-hybrid assays, mammalian two-hybrid assays score interactions between proteins that are localized in the nucleus (Fig. 1). Many transcription factors contain two functional modular domains: a DNA-binding domain (DBD) and a transcription activation domain (TAD). Whereas these domains are normally present in a single protein, their indirect interaction through protein-protein interactions can activate transcription as well (3). Thus, like an intact transcription factor, tethering of these two domains in a protein complex can promote transcription by interaction of the TAD with the basal transcription machinery. Typically, in two-hybrid assays, one protein is fused to the DBD and another protein of interest is fused to the TAD, and the direct or indirect interaction through another protein (or proteins) then activates transcription of a reporter gene under the control of a DBD binding promoter. Quantitation of the reporter expression then allows measurement of the protein interaction. Several combinations of DBDs and TADs have been successfully used in mammalian two-hybrid assays.Fig. 1.Schematic representation of the mammalian two-hybrid system. The Gal4 DBD, fused to protein X, binds to the Gal4 DNA sequence but does not activate transcription of the luciferase reporter, because it does not have a transcriptional activation domain or interact with one. The VP16 TAD, fused to protein Y, also does not activate transcription, because it lacks DNA binding. It is only when proteins X and Y interact with each other that the DBD is tethered to the transactivation domain, thus activating transcription of the luciferase gene.
 Fig. 1.
Fig. 1. Fig. 2.
Fig. 2. Fig. 3.
Fig. 3. Fig. 4.
Fig. 4. Fig. 5.
Fig. 5. Fig. 6.
Fig. 6.![]()
![]()
![]()
![]()
Gemma S. Puts & Natasha Spadafora , 2016, Springer Protocols
![]()
Gemma S. Puts & Natasha Spadafora , 2016, Springer Protocols
![]()
Shunwen Lu , 2012, Springer Protocols
Mammalian two-hybrid assays are used to analyze protein-protein interactions in transiently transfected mammalian cells. The concepts and principles that form the basis for mammalian two-hybrid assays are essentially the same as for the yeast
Mammalian two-hybrid assays are used to analyze protein-protein interactions in transiently transfected mammalian cells. The concepts and principles that form the basis for mammalian two-hybrid assays are essentially the same as for the yeast two-hybrid assays (1), discussed in other chapters, except that the analyses are carried out in transfected mammalian cells (2). Thus, similar to yeast two-hybrid assays, mammalian two-hybrid assays score interactions between proteins that are localized in the nucleus (Fig. 1). Many transcription factors contain two functional modular domains: a DNA-binding domain (DBD) and a transcription activation domain (TAD). Whereas these domains are normally present in a single protein, their indirect interaction through protein-protein interactions can activate transcription as well (3). Thus, like an intact transcription factor, tethering of these two domains in a protein complex can promote transcription by interaction of the TAD with the basal transcription machinery. Typically, in two-hybrid assays, one protein is fused to the DBD and another protein of interest is fused to the TAD, and the direct or indirect interaction through another protein (or proteins) then activates transcription of a reporter gene under the control of a DBD binding promoter. Quantitation of the reporter expression then allows measurement of the protein interaction. Several combinations of DBDs and TADs have been successfully used in mammalian two-hybrid assays.Fig. 1.Schematic representation of the mammalian two-hybrid system. The Gal4 DBD, fused to protein X, binds to the Gal4 DNA sequence but does not activate transcription of the luciferase reporter, because it does not have a transcriptional activation domain or interact with one. The VP16 TAD, fused to protein Y, also does not activate transcription, because it lacks DNA binding. It is only when proteins X and Y interact with each other that the DBD is tethered to the transactivation domain, thus activating transcription of the luciferase gene.